Creative Proteomics has rich experience in quantitative proteomics analysis. With a professional
team and advanced mass spectrometry facilities, we can provide you with
a one-stop stable isotope labeling by amino acids in cell culture (SILAC)-based
proteomics analysis service to accelerate your research progress.
The experimental principle of SILAC is to add the essential amino acids
lysine (Lys) and arginine (Arg) labeled with natural isotopes (light) or
stable isotopes (neutral or heavy) to the cell culture medium. After 5-6
doubling cycles, the stable isotope-labeled amino acid is completely incorporated
into the newly synthesized protein of the cell to replace the original
amino acid, so that it is labeled with a stable non-radioactive isotope.
Then, by mixing different types of proteins in equal amounts, the proteins
are digested into small peptide fragments using protease, and finally,
mass spectrometry analysis is performed. SILAC-based proteome analysis
has high analytical precision and breadth and is widely used in quantitative
proteomic analysis research.
Service Content of SILAC-Based Proteomics Analysis
In the SILAC-based proteomics analysis service, Creative Proteomics utilizes
stable isotope labeling under cell culture conditions to process cell samples,
which are then analyzed by mass spectrometry facilities. In this service,
we can provide a series of services including but not limited to cell culture,
cell labeling, protein extraction, proteolytic digestion, peptide separation,
mass spectrometry analysis, raw data analysis, and bioinformatics analysis,
etc. The SILAC-based proteomics analysis service can compare the area of
unlabeled/isotope-labeled peaks in the primary mass spectrum, to perform
relative quantitative analysis of proteins, and can also use the secondary
spectrum to sequence peptides assay to complete protein identification.
Mass Spectrometry Facilities and Projects
-
Liquid Chromatography-Mass Spectrometry (LC-MS)
-
Thermo Scientific™ Orbitrap Fusion™ Tribrid™ Mass Spectrometer
-
Thermo Scientific™ Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer
-
Thermo Scientific™ Q Exactive™ HF Hybrid Quadrupole-Orbitrap Mass Spectrometer
Requirements for Samples
-
Cell samples: cells that have been subcultured at least 5 times (both
cryopreserved samples and fresh samples); or samples that are self-labeled
cells and completed the treatment process such as stimulating cells.
-
If performing CEX fractionation, the recommended sample size is 50 to
100 μg.
-
0.5 to 5 μg is recommended for a single LC-MS/MS run.
-
Please provide the specific concentration, volume, preparation time, and
source of each sample. And inform the sample information as well as the
control and experimental samples (if there is a group, describe the group
information in detail.)
Deliverables
-
Experimental steps
-
Relevant mass spectrometry parameters
-
Details of the identified phosphorylation sites
-
Mass spectrometry images
-
Raw data
Our Advantages
-
Provide personalized experimental design to meet all your needs.
-
Professional technical appraisal team to ensure fast experimental cycle
and accurate experimental results.
-
Ultra-high protein labeling efficiency and the entire service process
is open and transparent, with no additional charges.
Service Process
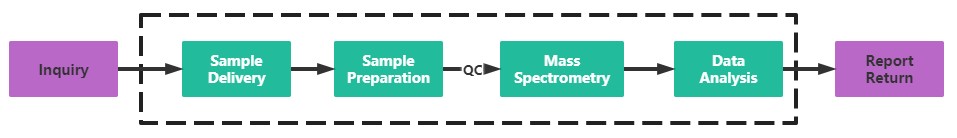
With professional mass spectrometry facilities and an experienced technical
team, Creative Proteomics can provide SILAC-based proteomics analysis service through mass spectrometry
facilities. If you are interested in our SILAC-based proteomics analysis
service, please
contact us
today for a solution.
The service is for research only, not for clinical use.