Creative Proteomics has an impressive track record in quantitative proteomics analysis. We
provide advanced high-throughput iTRAQ-based proteomics analysis service
to provide technical support to those engaged in protein research.
iTRAQ (isobaric tags for relative and absolute quantitation) uses multiple
isobaric tags for relative and absolute quantitation of proteins. This
is a patented technology for proteome quantification developed by AB SCIEX
in the United States. It can be applied to protein identification and quantification
in a variety of samples such as bacteria, yeast, human tissue, cells, and
body fluids, and can simultaneously compare the expression levels between
2 to 8 protein samples. As a new quantitative proteomics research method,
this technology exhibits high throughput, good reproducibility, and high
sensitivity, and has been widely used to study the pathogenesis of many
infectious pathogens.
Service Content of iTRAQ-Based Proteomics Analysis
In the iTRAQ-based proteomics analysis service, Creative Proteomics utilizes
isobaric reagents to label primary amines of peptides and proteins. The
role of the balancing group in iTRAQ reagents is to make the labeled peptide
in each sample isobaric and to facilitate quantification by analyzing the
reporter group generated upon fragmentation in the mass spectrometer. This
greatly facilitates peptide identification due to the higher intensities
of the precursor and fragment ions. Identify and quantify peptides in the
same experiment using multidimensional LC and MS/MS by comparing four MS/MS
reporter ions (range 114 to 117 Da) or eight MS/MS reporter ions (range
113-119 and 121 Da) peak area and the resulting peak ratio to obtain mass
spectrometry results. In this service, we can provide sample preparation
services such as cell lysis, trypsin hydrolysis of proteins to produce
tryptic peptides, etc. Bioinformatics analysis services such as functional
annotation and cluster analysis can also be provided.
Mass Spectrometry Facilities and Projects
-
AB SCIEX Triple Quad™ 6500 Mass Spectrometer
-
AB SCIEX QTRAP 5500 Ion Trap Mass Spectrometer
-
AB Sciex API 3000 ™ LC/MS/MS Triple Quadrupole Tandem Mass Spectrometer
Requirements for Samples
-
Samples must be prepared in buffers that do not contain primary amines.
-
The recommended sample size is 25 µg (8 plex) to 100 µg (4 plex).
-
Quantify at the MS/MS spectral level, typically 400 or more identified
proteins from the crude cell or tissue extracts.
-
Please provide the specific concentration, volume, preparation time, and
source of each sample. And inform the sample information as well as the
control and experimental samples (if there is a group, describe the group
information in detail.)
Deliverables
-
Experimental steps
-
Relevant mass spectrometry parameters
-
Details of the identified phosphorylation sites
-
Mass spectrometry images
-
Raw data
Our Advantages
-
The labeling efficiency is high, and almost all proteins in the sample
can be labeled.
-
Mass spectrometry has high detection sensitivity and can detect low-abundance
proteins.
-
Accurate quantification of multiple protein mixtures or recombinant protein
mixtures is possible.
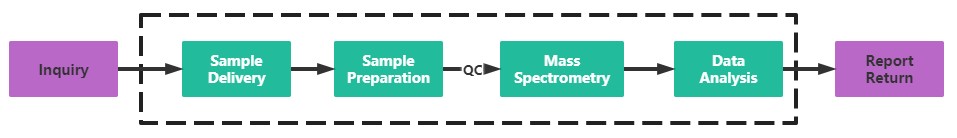
Service Process
With advanced mass spectrometry facilities and an experienced team, Creative Proteomics can provide researchers with professional protein quantitative analysis
services. If you are interested in our iTRAQ-based proteomics analysis
service, please
contact us
directly for details and quotation.
The service is for research only, not for clinical use.