Creative Proteomics has extensive experience in protein post-translational modification identification. With our professional team and advanced mass spectrometry facilities, we can provide you with one-stop protein citrullination identification services from experimental design to result in delivery, accelerating your research progress.
Citrullination of proteins is a post-translational modification process in which arginine residues in protein-peptide chains are converted into citrulline residues by protein arginine deiminases (PADs) in the presence of calcium ions. Citrullination is an important post-translational modification of proteins, which is involved in the pathogenesis of various diseases in vivo and has important pathological and physiological significance. Citrullination plays an important role in cancer, Alzheimer's disease, multiple sclerosis, and various autoimmune diseases, and is closely related to the occurrence and progression of the disease. Moreover, citrullination of histone arginine is one of the important epigenetic marks and is crucial for regulating chromatin remodeling.
Service Content of Protein Citrullination Identification
In the protein citrullination identification service, Creative Proteomics uses our advanced mass spectrometry facilities to identify citrullinated proteins and modification sites. The identification of citrullination is difficult, but we are confident that we can serve our customers to their satisfaction. In this service, we can use the monoisotopic mass increase (+0.984016 Da) to detect the citrullination of arginine by MS. Citrullination can also be detected using the monoisotopic mass reduction (-43.0058 Da) following dissociation fragmentation of the citrulline residue (HNCO) in isocyanate mass spectrometry. Citrullinated polypeptides can also be selectively labeled with 4-bromophenylglyoxal and then detected for citrullination of the protein by MALDI-MS analysis. For the analysis of complex protein and peptide mixtures, we pooled biotin with phenylglyoxal, producing biotin-PG (BPG) for streptavidin conjugation. After binding, it was digested by protease, and finally citrullinated by MS.
Mass Spectrometry Facilities and Projects
- Thermo Scientific™ Orbitrap Fusion™ Tribrid™ Mass Spectrometer
- Thermo Scientific™ Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer
- Thermo Scientific™ Q Exactive™ HF Hybrid Quadrupole-Orbitrap Mass Spectrometer
- Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS)
Requirements for Samples
- Solution (target protein): total target protein > 50 μg, target protein concentration > 80%.
- Solution (large-scale mixed protein): total protein > 1 mg, protein concentration > 1 μg/μL.
- Please store all samples at -80°C, do not freeze and thaw samples repeatedly, and use sufficient dry ice for transportation.
- Please provide the specific concentration, volume, preparation time, and source of each sample. And inform the sample information as well as the control and experimental samples (if there is a group, describe the group information in detail.)
Deliverables
- Experimental steps
- Relevant mass spectrometry parameters
- Details of the identified phosphorylation sites
- Mass spectrometry images
- Raw data
Our Advantages
- Efficient and professional protein citrullination identification service.
- Capable of analyzing a variety of eukaryotic and prokaryotic samples.
- It provides a basis for the occurrence, development, and early diagnosis of various diseases.
Service Process
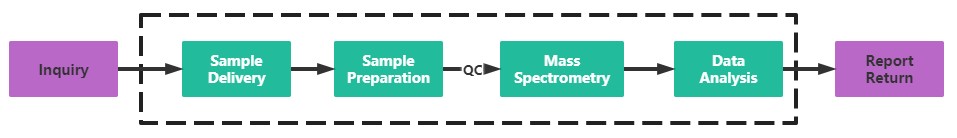
Creative Proteomics has built a mature mass spectrometry detection platform, and our professional mass spectrometry identification team can quickly and accurately solve the problem of protein citrullination identification for you. If you are interested in our protein citrullination identification service, please contact us today for technical support.
References
- Creese, A. J.; et al. On-line liquid chromatography neutral loss-triggered electron transfer dissociationmass spectrometry for the targeted analysis of citrullinated peptides. Anal Methods. 2011, 3(2): 259-266.
- Hao, G.; et al. Neutral loss of isocyanic acid in peptide CID spectra: a novel diagnostic marker for mass spectrometric identification of protein citrullination. J Am Soc Mass Spectrom. 2009, 20(4): 723-727.
- Clancy, K. W.; et al. Detection and identification of protein citrullination in complex biological systems. Curr Opin Chem Biol. 2016, 30: 1-6.
The service is for research only, not for clinical use.