Creative Proteomics has extensive experience in proteomic analysis and provides you with high-resolution
protein-protein interaction (PPI) identification service with a well-established
and advanced technology platform.
Protein-protein interactions are highly specific physical contacts between
protein molecules due to interactions such as hydrogen bonding. Proteins
can hardly function by themselves in organisms, and many physiological
processes rely on protein-protein interactions to function. To identify
protein interactions is to identify proteins that have interacting relationships.
Using mass spectrometry identification technology, identify the protein
that can interact with a specific protein in the experiment, to determine
the interacting protein of a specific protein. This is of great significance
for revealing the functional structure of proteins, deeply understanding
the physiological processes involved in proteins, and discovering protein
targets.
Service Content of Protein-Protein Interactions Identification
In the mass spectrometry-based protein interaction identification service
provided by Creative Proteomics, co-immunoprecipitation (CO-IP), GST fusion
protein Pull-down technology, and SILAC combined with mass spectrometry
are usually used for analysis. These methods have their advantages. The
co-immunoprecipitation method is mainly to separate and identify the interacting
protein complexes in the natural state; the GST fusion protein Pull-down
technology can eliminate intracellular interference. In our protein interaction
experiments, we first separated the interacting proteins and then used
mass spectrometry to identify and analyze the obtained protein of interest/interacting
protein. For the identification of known proteins that do not require high
accuracy, primary mass spectrometry can complete the identification by
obtaining peptide mass fingerprints; if the accuracy is extremely high
or for unknown proteins, tandem mass spectrometry can be used for identification.
Mass Spectrometry Facilities and Projects
-
Thermo Scientific™ Orbitrap Fusion™ Tribrid™ Mass Spectrometer
-
Thermo Scientific™ Orbitrap Fusion™ Lumos™ Tribrid™ Mass Spectrometer
-
Thermo Scientific™ Q Exactive™ HF Hybrid Quadrupole-Orbitrap Mass Spectrometer
Requirements for Samples
-
Please confirm that the total protein is greater than 20 µg, and the solution
and lyophilized powder are acceptable.
-
Please provide the specific concentration, volume, preparation time, and
source of each sample. And inform the sample information as well as the
control and experimental samples (if there is a group, describe the group
information in detail.)
Deliverables
-
Experimental steps
-
Relevant mass spectrometry parameters
-
Details of the identified phosphorylation sites
-
Mass spectrometry images
-
Raw data
Applications of Protein-Protein Interactions Identification
-
Explore the mechanisms of disease development.
-
Determination of large-scale protein interaction network mapping.
-
Finding new drug targets, developing new drugs, and pioneering new ways
to treat diseases.
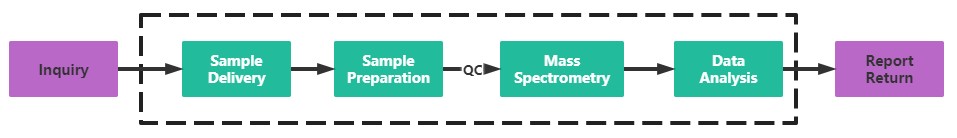
Service Process
Creative Proteomics is an expert in protein interaction identification, and we can provide
an extremely accurate protein-protein interactions identification service.
If you are interested in our protein interaction identification service,
don't hesitate to contact us today for a quote.
The service is for research only, not for clinical use.